1: Mapping cellular connections across large portions of the brain is challenging. Historically, it has required massive expense with electron microscopy, or sacrificing ultrastructural resolution for volumetric throughput with light microscopy.
Expansion microscopy — chemically expanding the tissue and then imaging it with light microscopy — has emerged since the mid-2010s as an option for improved resolution without the high cost of electron microscopy.
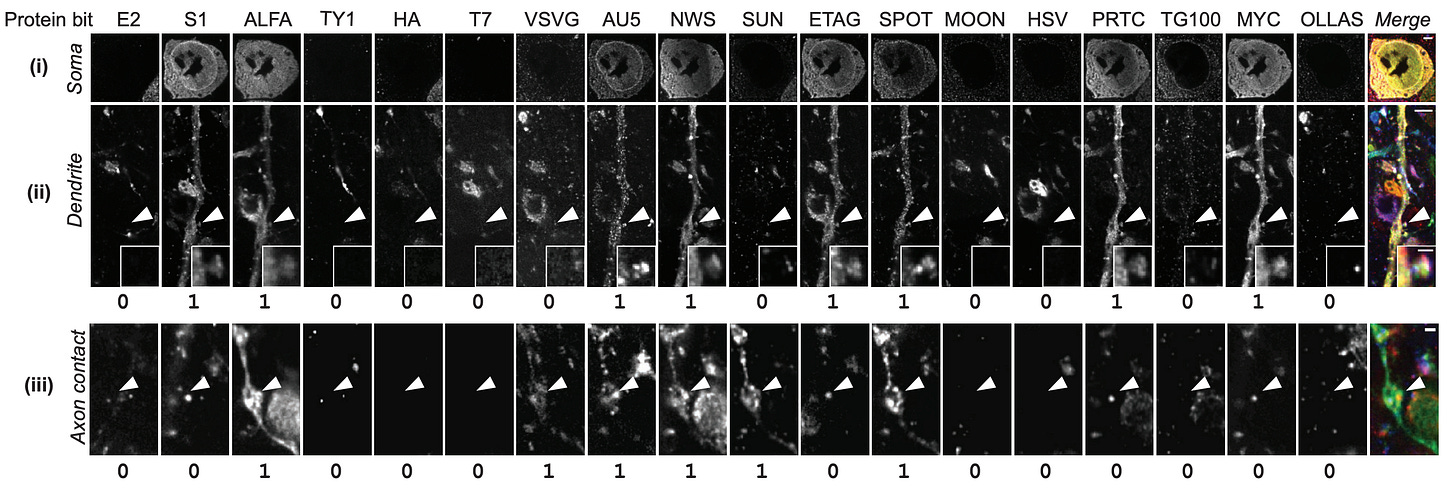
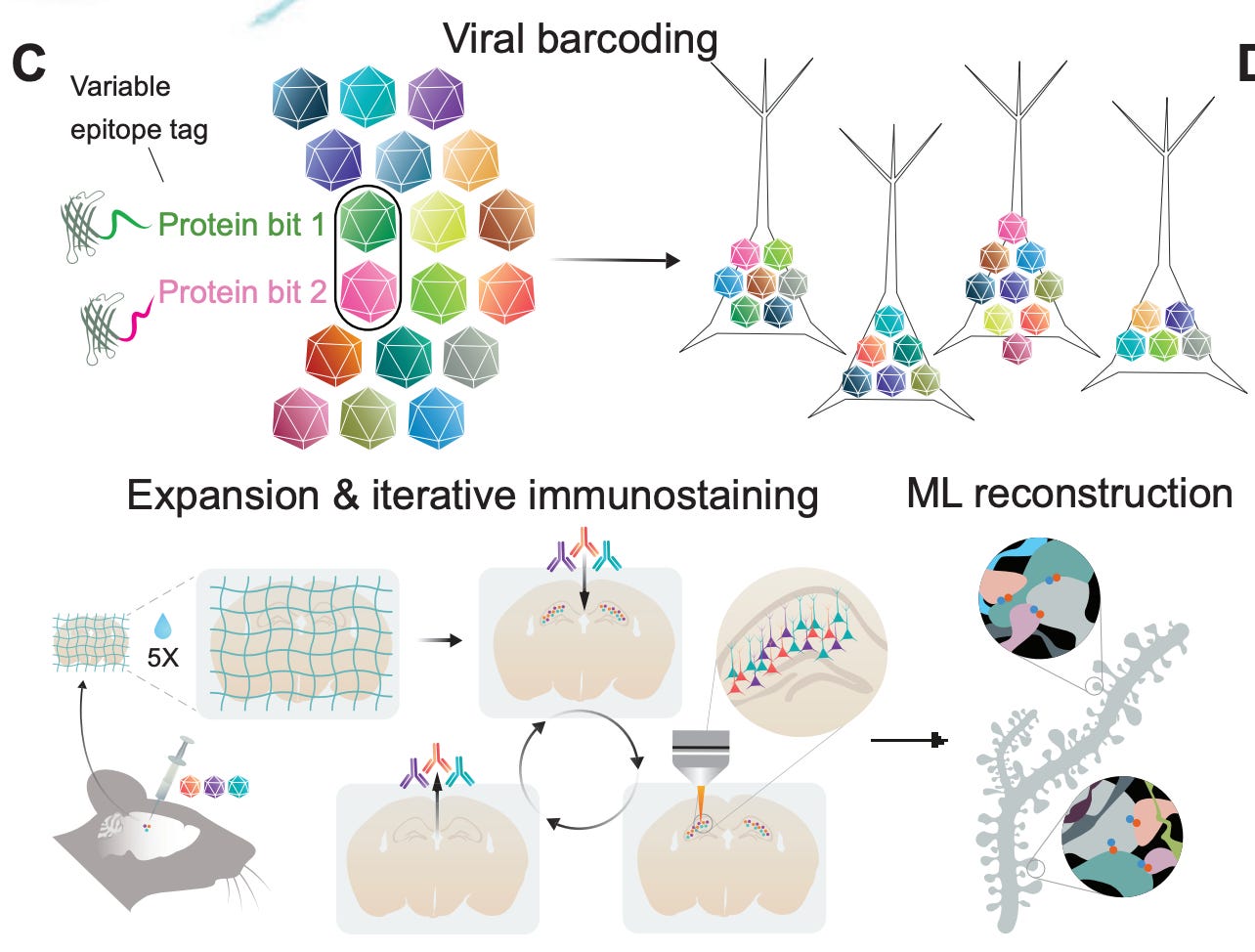
A fascinating new paper from the team at E11 Bio introduces a method to combine expansion microscopy with a clever molecular barcoding system to map the brain. They inject viral vectors (AAVs) carrying 18 different protein “bits” — i.e., variants of GFP tagged with distinct epitopes — that cells then take up and randomly express in different combinations:
As a result, each neuron that takes up the virus gets a unique molecular barcode (theoretically up to 262,143 combinations), which is expressed throughout the cell.
To preserve the mouse brains, they perfused them with formaldehyde, removed them from the skull, and post-fixed them under refrigeration for 28 hours. After using the Magnify expansion method and other forms of processing, the barcodes can then be read out for iterative rounds of immunostaining and imaging:
One limitation with their current barcoding approach is that it only labels a subset of neurons. In the hippocampal dataset, only 49% of cell bodies (146 out of 298 somas) expressed at least one protein bit. Among those labeled cells, 80% had unique barcodes, so one in five labeled neurons also shared its barcode with at least one other cell.
This sparsity is due to their stochastic AAV infection approach. Each neuron independently takes up some random subset of the 18 different viruses in the pool, leading to combinatorial diversity but incomplete coverage.
This makes their approach different from electron microscopy or dense light microscopy methods, which aim to reconstruct every neuron in a volume. As it currently stands, their method is better suited for applications requiring partial reconstruction, rather than building complete wiring diagrams.
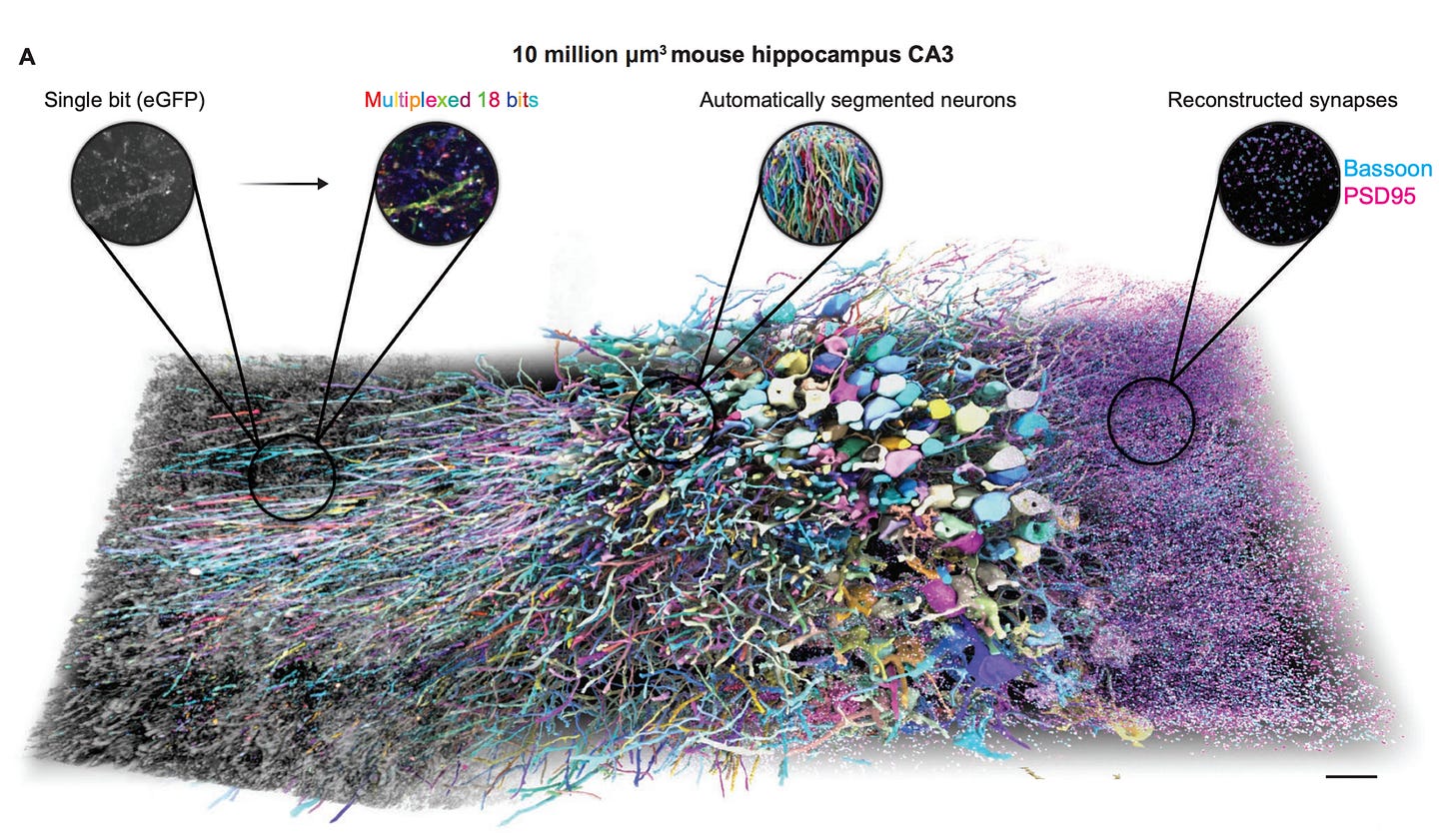
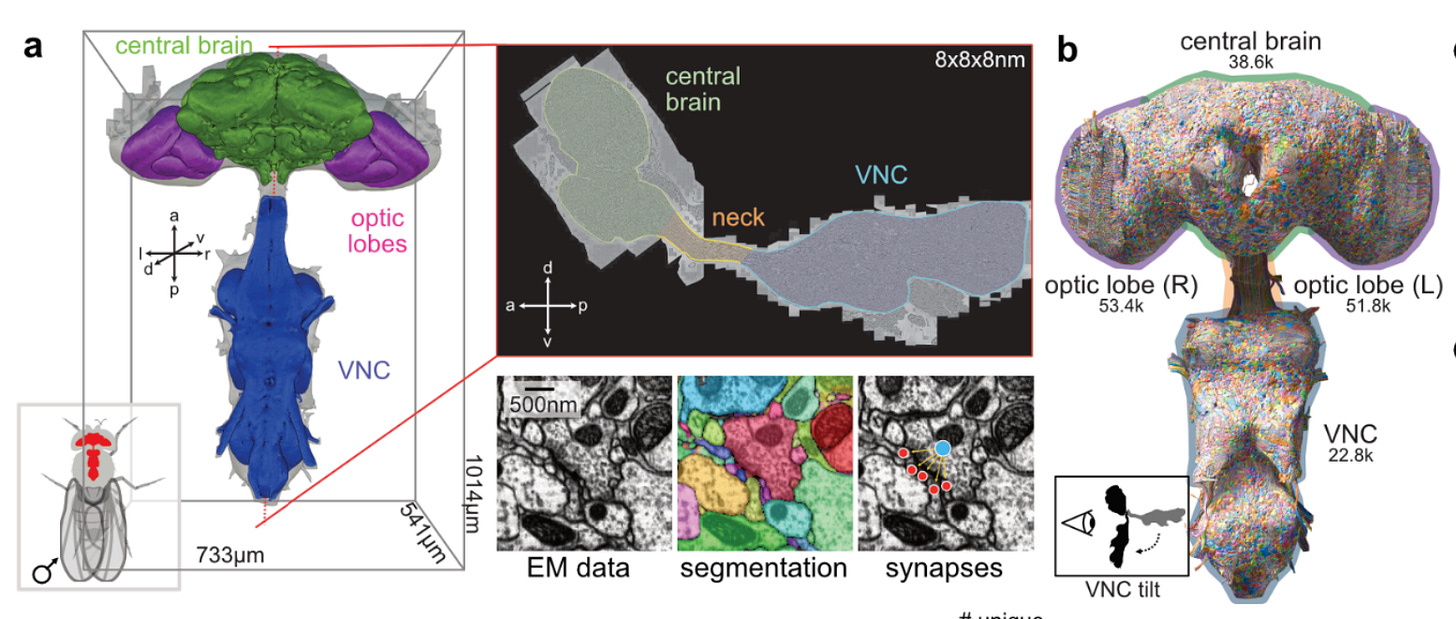
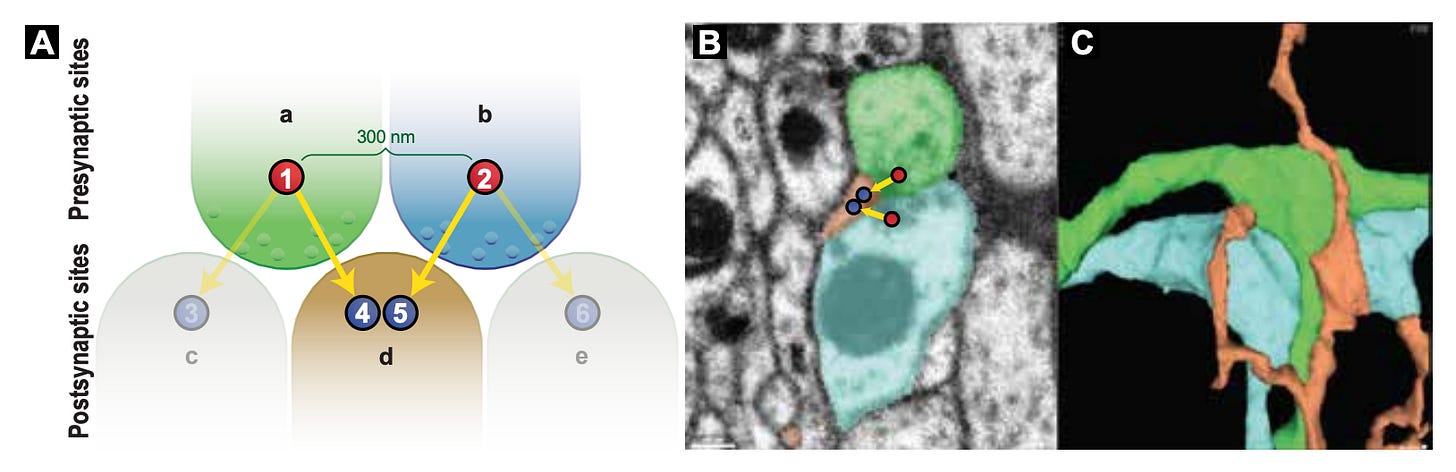
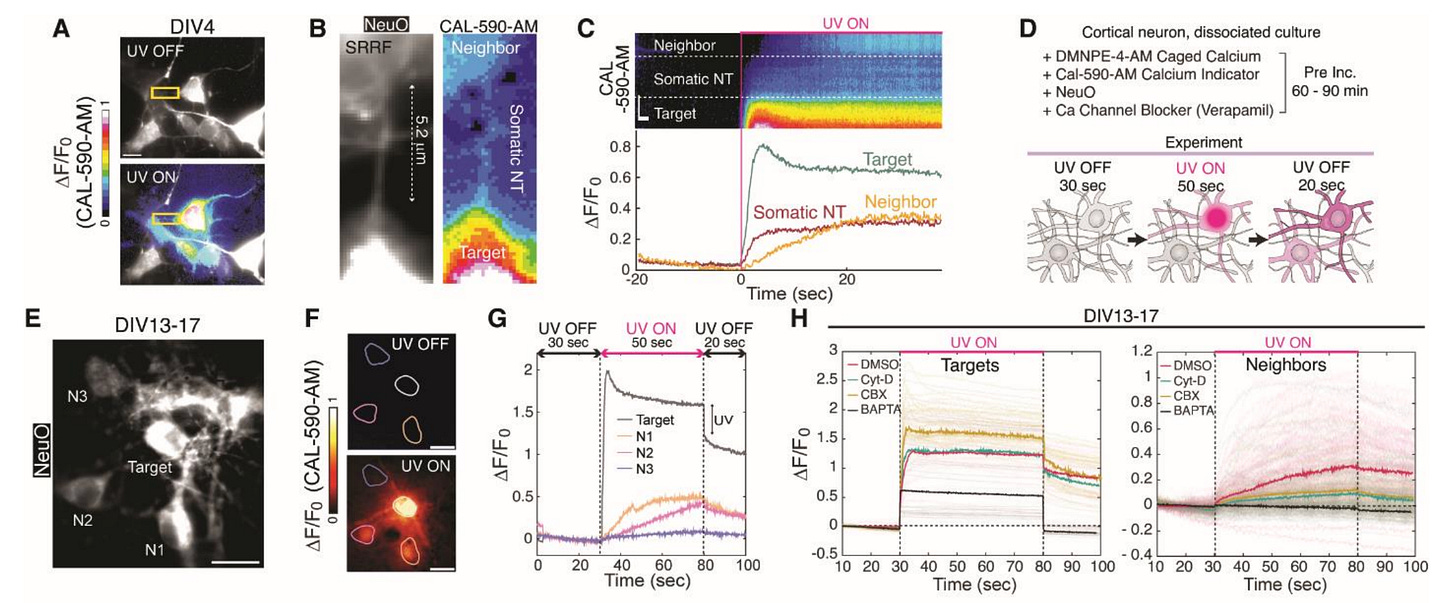
Using this approach on 5-fold expanded mouse brain tissue, they were able to reconstruct individual neurons across a 10 million μm³ volume of the hippocampus, with each neuron assigned a different pseudo-color based on its unique barcode. This led to a map around 9 million synapses throughout the tissue, which were identified by the co-localization of presynaptic (Bassoon, cyan) and postsynaptic (PSD95, magenta) markers:
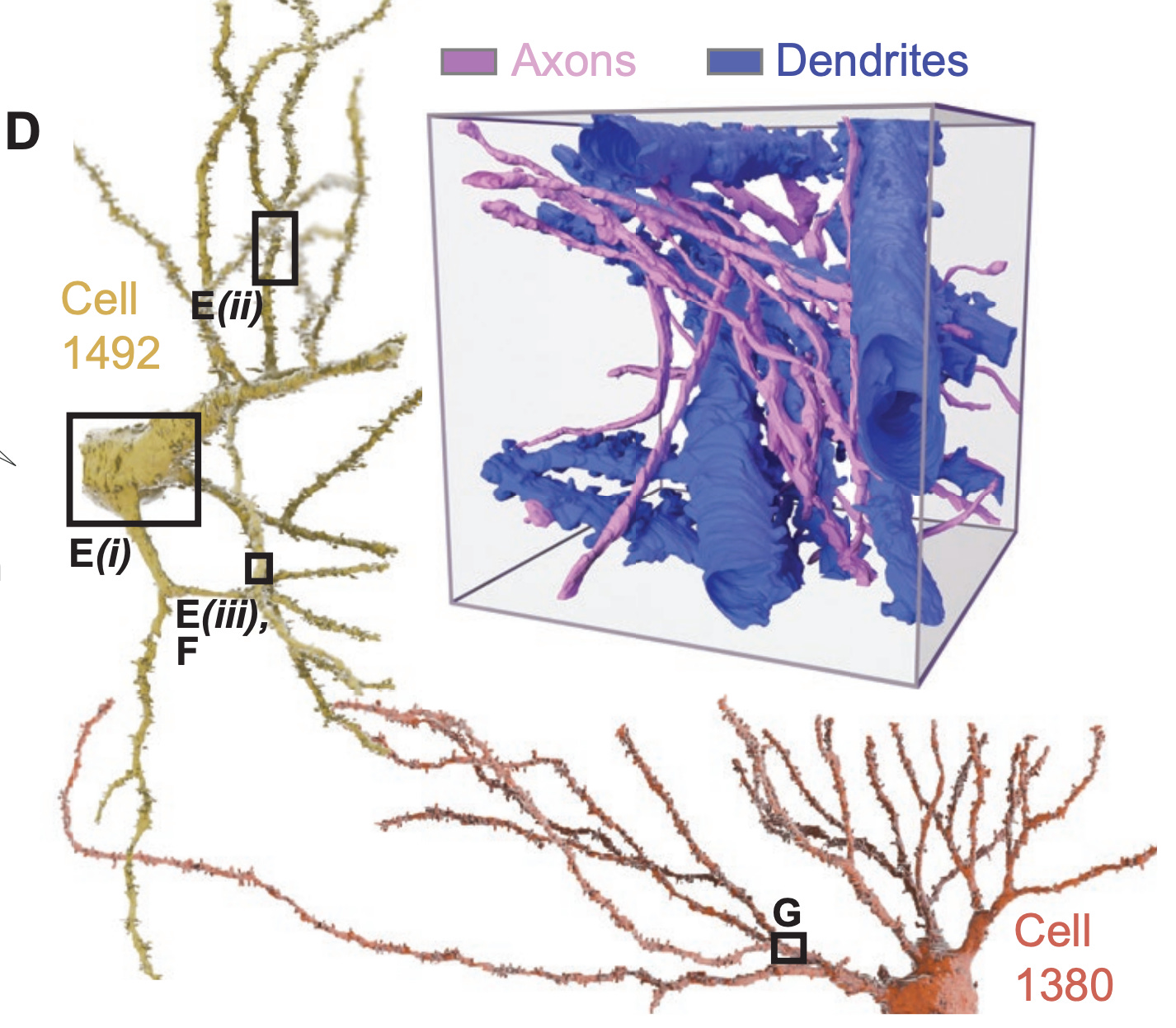
Here is an example of two reconstructed neurons with their morphologies traced. The inset displays a higher-resolution view of neuronal processes, with the axons and dendrites labeled to identify putative synaptic connections. Note that axons tend to be thinner than dendrites, which is also seen here:
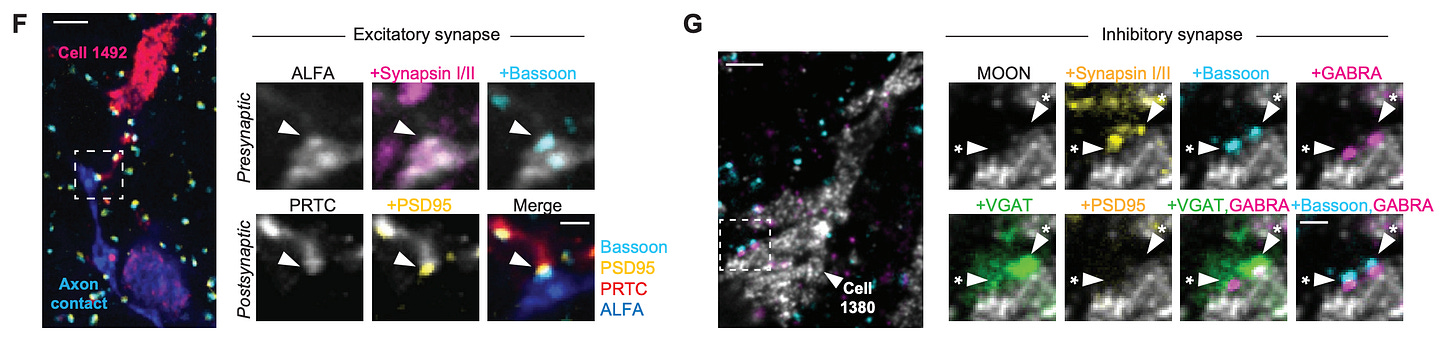
Because they use light microscopy, they are also able to relatively easily stain for particular biomolecules, which is a huge advantage over electron microscopy, where this is possible but more difficult. Here they show staining for different biomolecules at synapses, which can be seen in highly localized sub-cellular positions:
The senior author Andrew Payne notes in an interview that the mission of the company is to eventually enable the mapping of the whole human brain.
Because they use viral vectors to express the barcodes, the options for human application seem relatively limited. Injecting viral vectors into living human brains purely for research purposes seems pretty much out of the question (?). And keeping large volumes of postmortem tissue viable long enough for the viral vectors to express and the barcodes to fill the cells seems difficult.
Another option would be to try to identify endogenous molecular barcodes, i.e. unique combinations of naturally expressed proteins, RNA transcripts, or other molecular markers that already distinguish individual neurons from each other to a sufficient degree of accuracy. If enough natural variation exists between cells, they might be able to use these innate molecular signatures for tracing without needing to deliver artificial barcodes.
One particularly promising candidate are protocadherin proteins, which appear to create locally unique expression patterns in each neuron, allowing neurons to distinguish self from non-self when making connections. The main problem is probably copy number, as protocadherins do not seem to be nearly as abundant as the proteins virally expressed in this paper. So other candidates or combinations would probably need to be identified.
I am especially interested in the endogenous molecular barcode option because it is one method I believe could be used to infer the original morphological states in the presence of damage following brain preservation. So I would really love to see this idea of mapping endogenous cellular barcodes explored more.
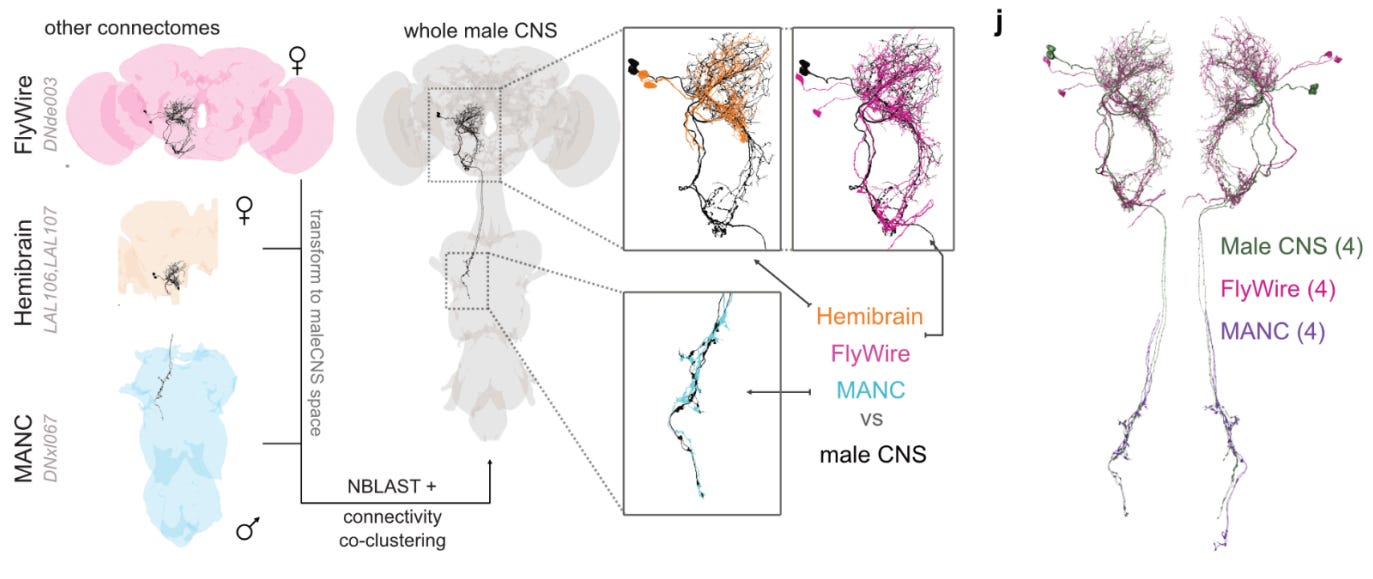
2: The complete connectome of an male entire Drosophila central nervous system has now been published. It contains 166,691 neurons.
Not sure how this fits into previous Drosophila connectome efforts? Don’t worry, the authors compare it to a few of those — one from a female brain (“Flywire”), one from a hemibrain, and one from a Male Adult Nerve Cord (MANC). All of these connectomes were highly similar. They were able to match 96.4% of central brain neurons to neurons in at least one of the previous connectomes.
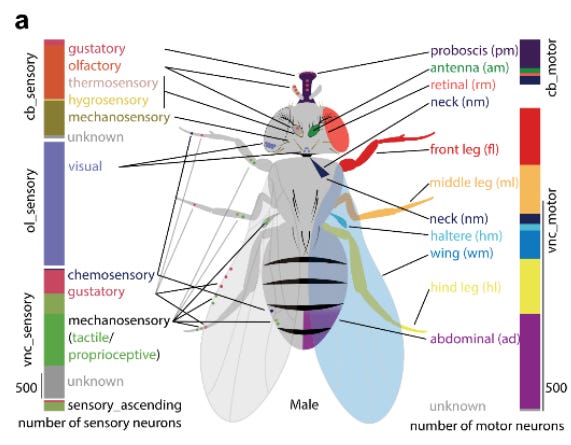
Interestingly, they were able to identify aspects of sensory neurons — both which nerve they enter through (antenna, leg, wing, etc.) and their modality (vision, taste, smell, touch, temperature, humidity, etc.) — based on anatomical knowledge of nerve organization, without needing to preserve the peripheral receptors or body parts themselves. Similarly, motor neurons could be catalogued based on their exit nerves and muscle targets inferred from anatomical knowledge. This is an example of how the essential information for brain functioning, from sensory processing to motor control, is contained within the CNS alone:
3: A new comparative connectomics study compares two Drosophila mushroom body data sets prepared and imaged separately by FIB-SEM. They found that the convergent synaptic motif, which is the name for a microcircuit in insects in which two nearby axons form presynaptic sites onto the same postsynaptic neuron, was present repeatedly across individuals. They suggest that that it may be a conserved building block of neural computation:
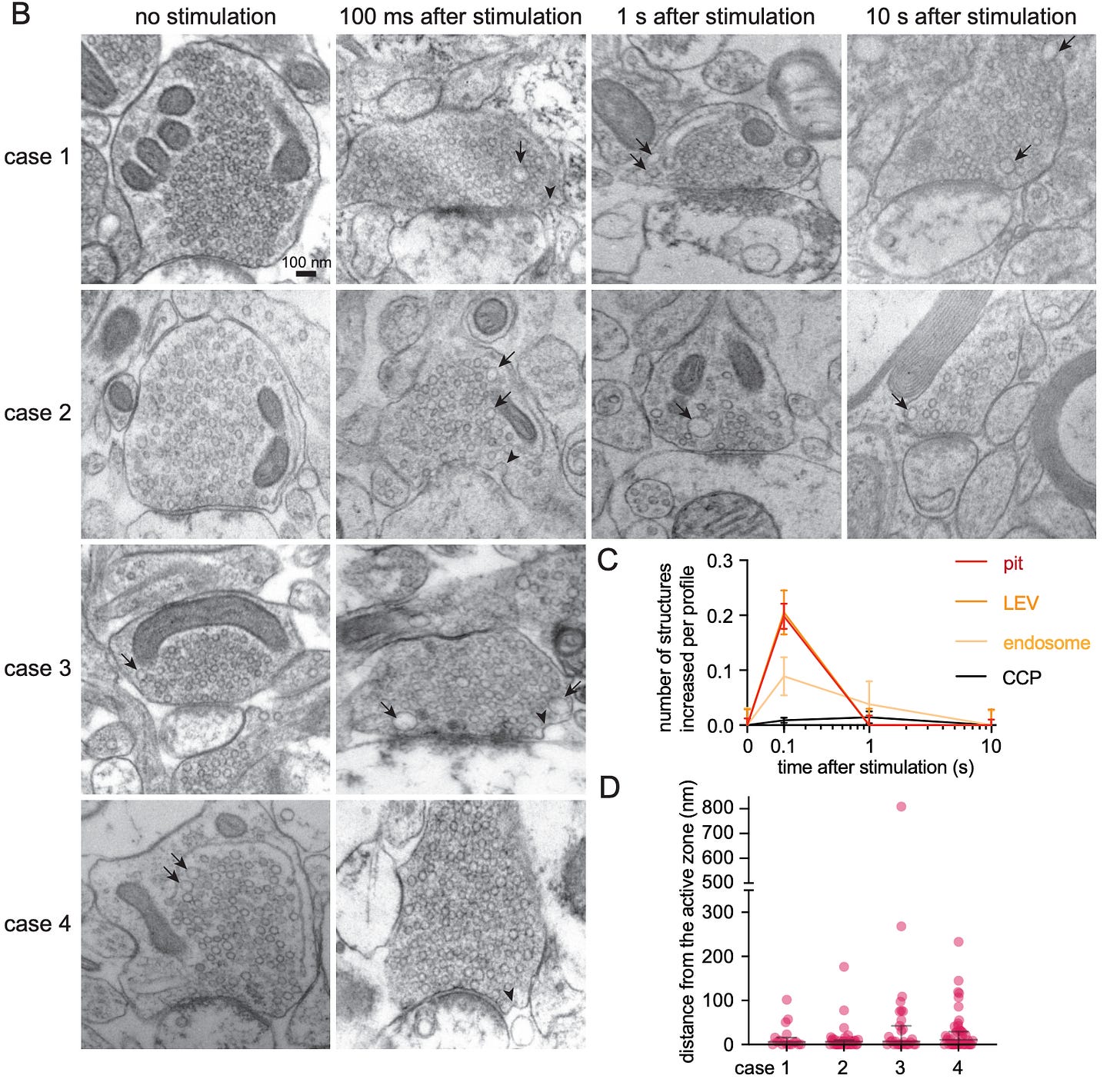
4: What happens to human synapses immediately after electrical stimulation? A new study uses acute brain slices from epilepsy surgery patients, i.e. freshly resected tissue that is kept viable after removal for a short period of time, to study this for the first time in humans. They find it is basically the same as in mice.
Before electric field stimulation, there are normal resting synapses with synaptic vesicles. 100 ms after stimulation, you can see uncoated endocytic pits (i.e. membrane invaginations that enable synaptic vesicle recycling, indicated by black arrowheads). By 1 second, you see mostly endosomes (i.e. membrane compartments, indicated by black arrows), which can remain for at least 10 seconds:
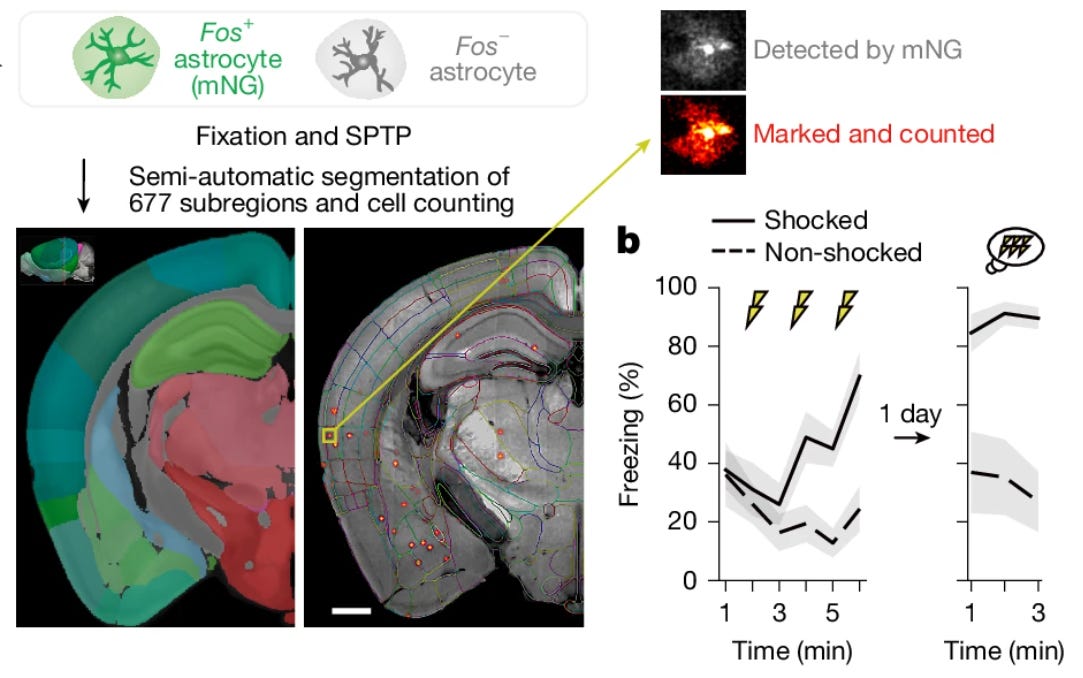
5: A new study uses whole-brain astrocyte Fos tagging in mice during a fear conditioning task. Fos is a protein commonly used as a measure of neural activity. They find that astrocytes play a role in providing memory stabilization after recall.
The process involves two steps. First, the initial memory formation upregulates adrenergic receptors over around 24 hours. Second, subsequent recall triggers these primed astrocytes to express Fos and secrete the protein IGFBP2, which they show stabilizes the memory. The authors were able to visualize these multi-day molecular traces in astrocytes following aldehyde fixation:
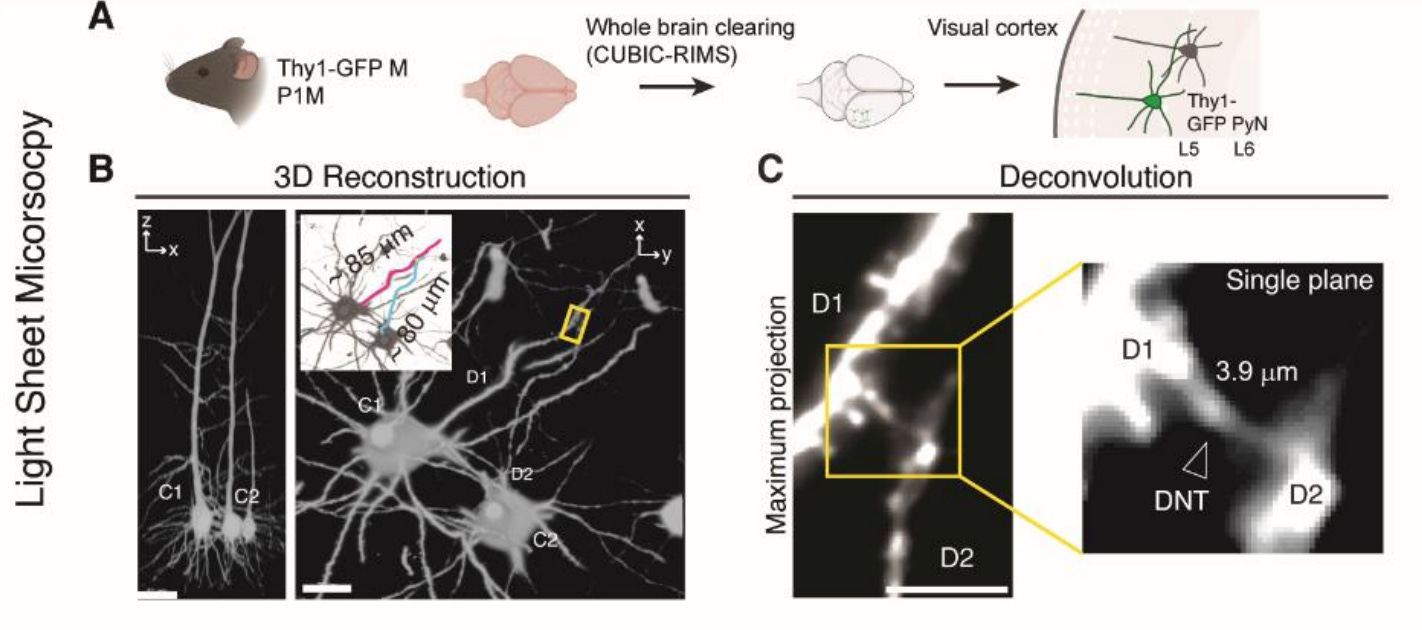
6: A new study characterizes a poorly understood form of direct communication between neurons called dendritic nanotubes. These are long, thin, actin-rich bridges that connect dendrites to one another directly rather than through synapses. They were able to visualize them after fixation and brain clearing:
They found that these nanotubes allow for the spread of ions and molecules between connected neurons. This enables them to propagate calcium signals and also to facilitate the spread of pathologic molecules such as amyloid-beta:
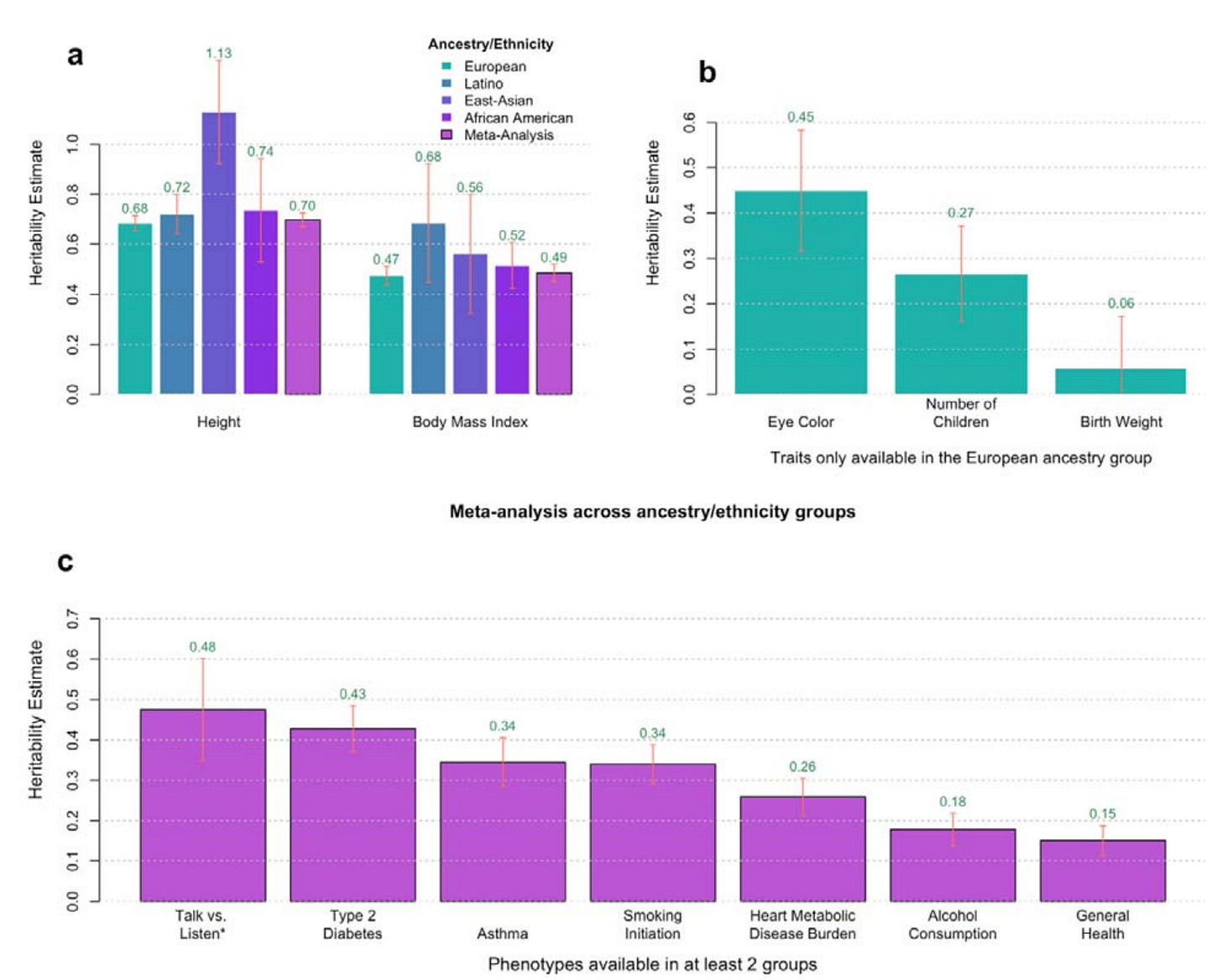
7: A new study uses genomic data from ~500,000 sibling pairs to estimate heritability for 14 traits. The key innovation is the use of a within-family design, which compares siblings who share the same family environment but randomly differ in how much DNA they inherited from each parent.
Traditional between-family studies compare unrelated individuals. The within-family design allows them to mitigate shared environment confounding (e.g. parenting, diet, socioeconomic status), avoid population stratification (i.e. ancestry-related environmental differences), and separate direct genetic effects from “genetic nurture” (i.e. the environments that parents create based on their own genes). They found some differences using this method. For example, they found that the heritability of type 2 diabetes was 0.43 (within-family) vs 0.62 (between-family), suggesting that previous heritability estimates were inflated by shared family lifestyle.
It’s an elegant design but requires a large sample size. Enter 23andMe. (It’s a general rule that most genomics data is not really that useful to a person on its own, but it only becomes useful when aggregated in very large data sets.)
Of the traits they studied, I found “Talk vs listen” to be the most interesting, as they found that it is very heritable (0.48) and I haven’t heard much about it or related traits before. It was based on a question that 23andMe users were asked: “If forced to choose, would you say you generally prefer to talk or to listen?” (0: Talk, 1: Listen, NA: I’m not sure). It was found to be more heritable than even type 2 diabetes or asthma.
I guess the takeaway is that the next time someone says that you are talking too much you can say it’s not your fault, it’s just your DNA.
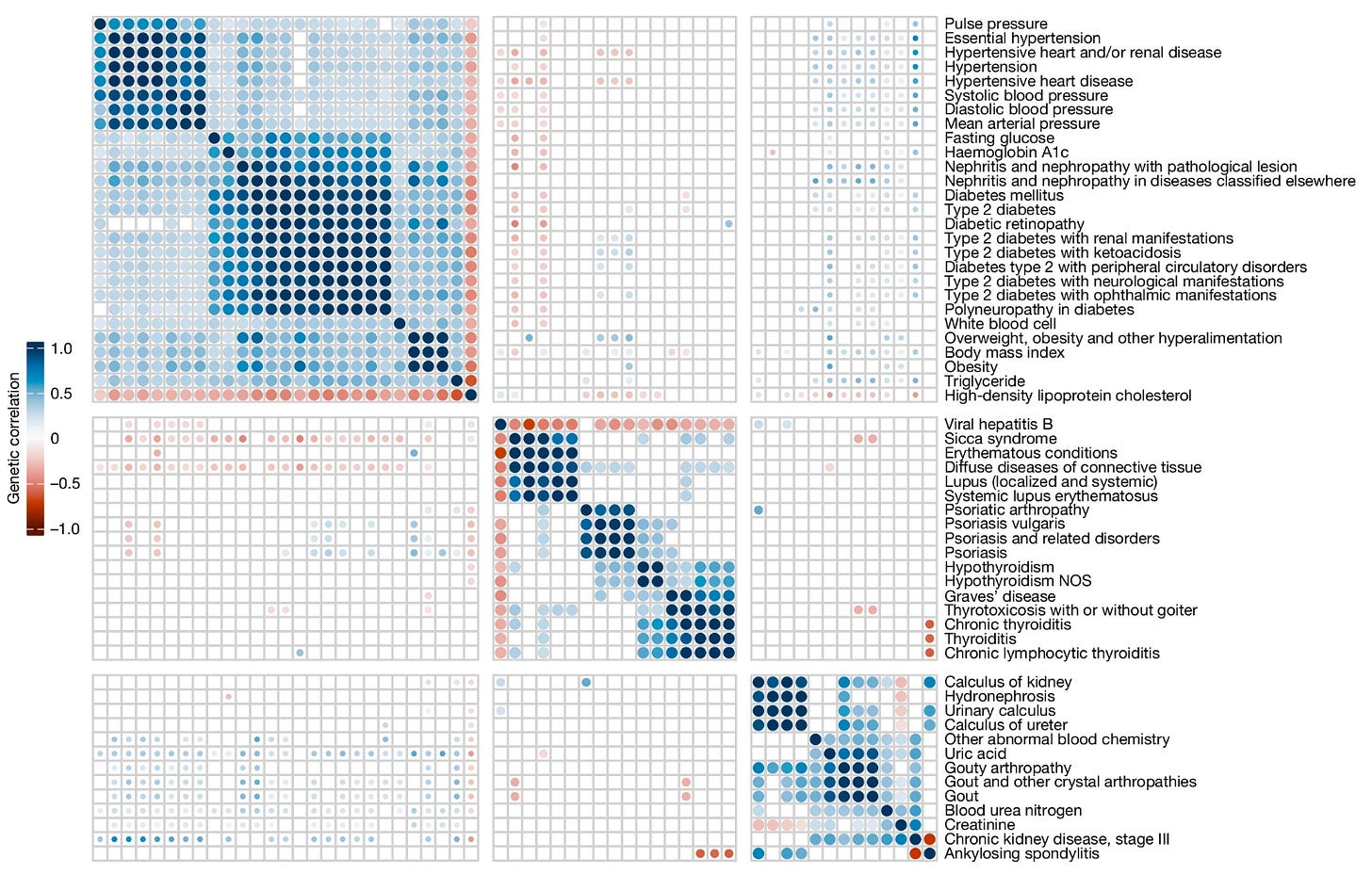
8: New genomics study from more than half a million Taiwanese residents of primarily Han Chinese ancestry with matched electronic medical record data.
I was particularly interested in the correlations among genetic risk scores they found for different phenotypes. In particular, they found that there were basically three genetically correlated clusters of conditions: (a) “metabolic syndrome” (e.g. obesity, high cholesterol, diabetes, high blood pressure), (b) “autoimmune diseases” (e.g. Sjögren’s syndrome, lupus, psoriasis, thyroid disease), and (c) kidney-associated conditions (e.g. chronic kidney disease, kidney stones, gout, ankylosing spondylitis):
9: Abhishaike Mahajan on the moral case for working in biology. My comment pointed out that while I fully agree with this, such exhortations must be balanced against the practical difficulties of sustaining a career in biomedical research. Plenty of people would like to work in biomedical research but find that they effectively cannot do so for reasons outside of their control.
10: Related: Career advice from Thomas Insel, a former director of the NIMH. He points out that a lot of scientific research these days (2025) is done at for-profit companies.
11: Randomized trial of the investigational new drug seltorexant for insomnia (n = 364, 2 week trial).
One of the key outcome metrics was WASO-6 — basically, the total minutes spent awake during during the first 6 hours after the person initially fell asleep. You want this to be lower (obviously). They found that seltorexant had better WASO-6 outcomes compared to placebo (around 30-40% improvement) and compared to zolpidem (Ambien) at 2 weeks (around 30% better), whereas zolpidem’s initial efficacy at day 1 waned.
Seltorexant is a selective orexin-2 receptor antagonist. Compared to dual orexin receptor antagonists (DORAs) that also antagonize orexin-1 receptors, they argue that the selective orexin-2 antagonism will help preserve non-REM sleep. It would have been nice to see a direct comparison with a DORA (a few of which have been approved), but it’s understandable they wouldn’t want to risk not seeing any improvements.
12: A capacity evaluation is a clinical assessment of a patient’s mental ability to make their own medical decisions. At least in the US, this is commonly thought of as an aspect of psychiatric expertise. There are existing frameworks on how to do capacity evaluations, but not as much guidance on whether to do them.
Friend of the blog Jacob Appel has written a new article with a three step guide for making that decision. First, to assume capacity exists without evaluation unless there is reasonable uncertainty. Second, to determine whether the results of the evaluation would impact patient care anyway. And finally, to weigh the potential benefits of the assessment against its costs in terms of the patient’s well-being.
13: Jan Betley reports having isolated anhedonia while taking semaglutide that improved upon decreasing the dose.
14: An argument that valproate (Depakote) should have REMS regulation for women of reproductive age due to its very high risk of teratogenicity.
15: Nolan Williams was famous in the psychiatry community for many reasons, including studies of accelerated TMS for depression (such as the SAINT trial). He recently died. Owen Scott Muir wrote a eulogy for him.
16: Tomás Bjartur wrote a post on the technical feasibility of molecular nanotechnology. Points out the difficulty of proving that it would not be possible:
“The design space is very large. Any proof that Drexlerian nanotechnology is impossible would have to be very robust. And it strikes me that most plausible looking arguments can likely be hacked around given how large the design space is.”
A good comment from SE Gyges: “[I]n every case people tend to think that things are possible only when they know more exactly how they will be done.”
17: Start-up Bexorg raises 23M for its technology using postmortem brain perfusion and profiling as a method for drug discovery.
18: Are you going to be in San Diego in mid November, for example going to the Society for Neuroscience (SfN) conference? If so, you might consider going to the Aspirational Neuroscience Awards event on November 17 at 7 pm PT at the Hilton San Diego Bayfront. Here are the previous award winners. Here is the previous panel discussion from 2023. To RSVP and learn more about this year’s event, see here.
19: On Nov 13th at 11 AM PT, I will be having an informal discussion with Aschwin de Wolf on the Cryosphere Discord on the differences between fixation and pure cryopreservation. Here is the invite link.
20: Charles Platt, author and former Director of Suspension Services at Alcor, came to visit us at Sparks Brain Preservation and wrote about our organization. Discussion on Reddit here and here. Here is his Substack post, which is quite interesting (at least to me) and has a few plot twists:
It was wonderful to meet Charles in person. We went to lunch in Salem. I ordered an Impossible burger and french fries. He pointed out that my meal was “not very life extensionist.” I was forced to concede the point. I could only nod gravely and continue chewing.
21: Posting on NN may be light for awhile as my wife and I just welcomed our second child. Thank you as always for your support of this newsletter.


For someone like me, not involved in the field but very interested in it, these monthly summaries are incredible.
Thank you so much for making them!
The work being done by Bexorg's team piqued my interest many years ago, and I was happy to see them raise more funding. Interestingly, both Bexorg and E11 Bio have a mutual philanthropic backer, though that might be unsurprising given the somewhat-overlapping interests of each company. Also, to echo other comments, a big congratulations to you and your wife!