The paradigm of differential network interactions
It is quite common in biology (and neuroscience, as a special case) for researchers to employ differential gene expression analysis, which produces lists of up- and down-regulated genes between a given set of conditions. And as Ideker and Krogan point out in their Jan '12 paper, this principle has already been extended to differential protein expression and post-translational modifications.
The authors go on to discuss how this approach has also been applied, with less fanfare, to differential interaction network analysis. In this paradigm, if an interaction between nodes (e.g., protein concentrations) in the network is present above noise in one condition, but not another, then they would call that a differential interaction.
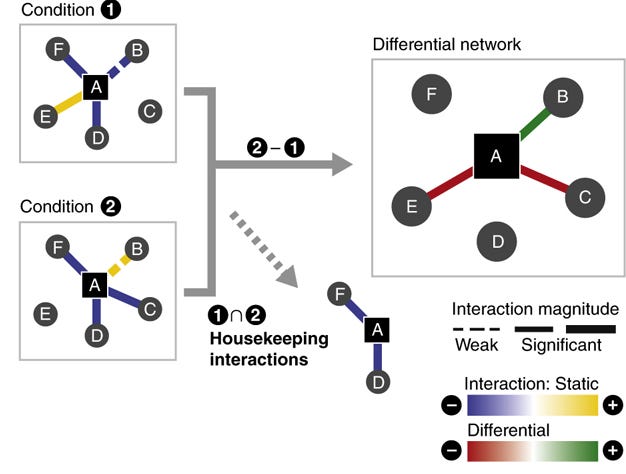
Very similar ideas can be applied to the study of neuronal network function. If we can say that an interaction between neuronal "nodes" (which could be, depending upon the scale, neurons, cortical columns, or brain regions) is differentially present between healthy and disordered states, then it suggests that that interaction is somehow involved with the disorder.
This is not a perfect paradigm, in part because the network "connections" can be less representative of the physical reality than we'd like, but I anticipate that we have much to mine from it about the operations of the nervous system.
Reference
Ideker T, Krogan NJ. 2012 Differential network biology. doi:10.1038/msb.2011.99
